Molecular Embeddings
The "Embed Molecule" feature in ChemXploreML is used to convert molecular structures from SMILES strings into numerical vectors, a process known as embedding. These embeddings are essential for machine learning tasks, as they provide a quantitative representation of the molecules.
Overview
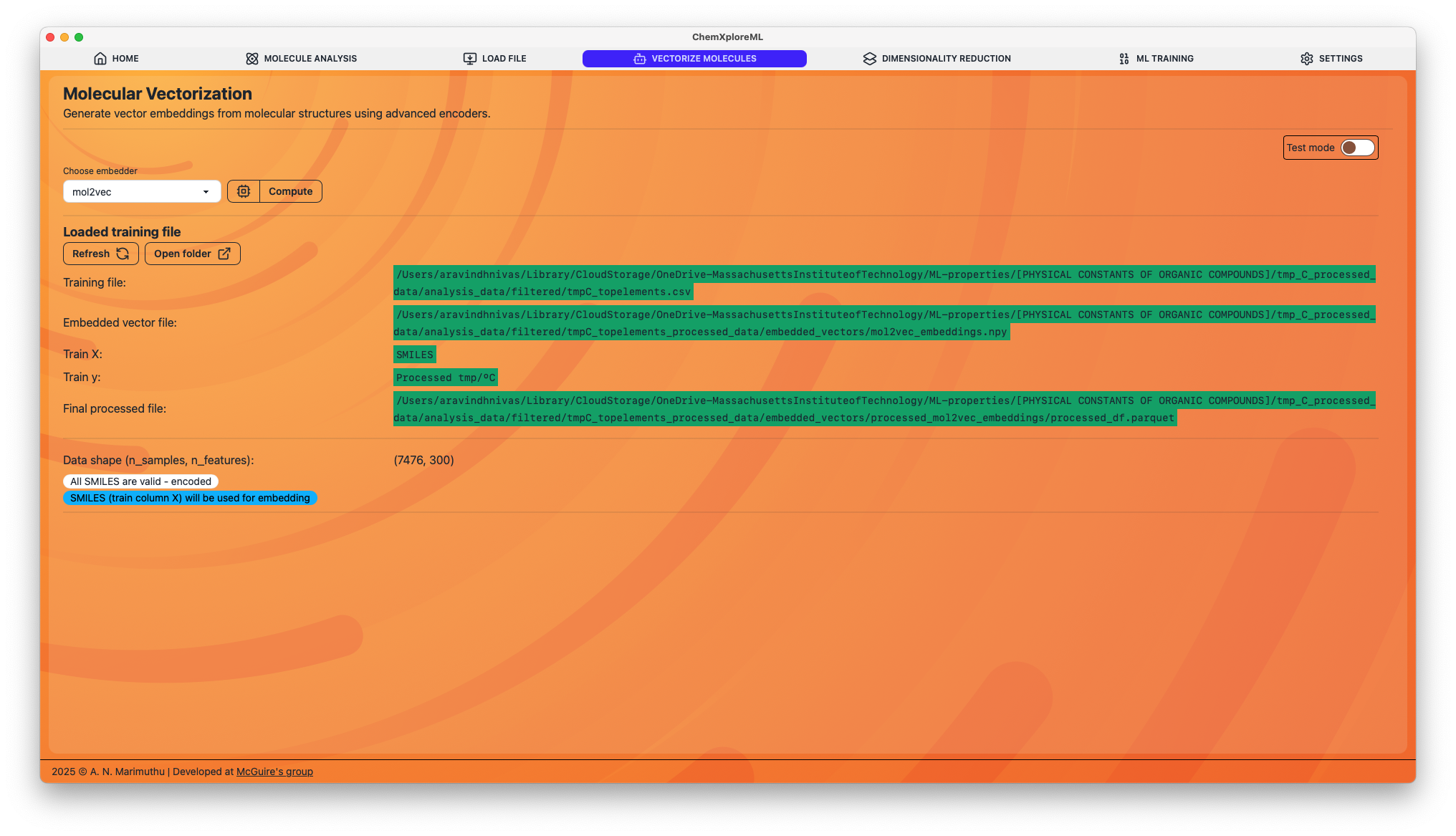
This module allows you to generate embeddings using several state-of-the-art models. You can either process an entire dataset ("File Mode") or test a single molecule ("Test Mode").
Supported Embedding Models
ChemXploreML supports the following pre-trained models for generating molecular embeddings:
- VICGAE: A graph-based model for learning molecular representations.
- mol2vec: An unsupervised machine learning approach to learn vector representations of molecular substructures.
- ChemBERTa-zinc-base-v1: A transformer-based model pre-trained on a large dataset of molecules.
- MoLFormer-XL-both-10pct: Another powerful transformer-based model for molecular embeddings.
For each of these models, ChemXploreML can automatically download the necessary pre-trained weights if they are not already available. You also have the option to use your own pre-trained model files.
Workflow
1. Test Mode
If you want to quickly see the embedding for a single molecule:
- Enable the Test mode checkbox.
- Enter a SMILES string in the input field.
- Select an embedder from the dropdown menu.
- Click the "Run" button.
- The resulting numerical vector will be displayed in the text area.
2. File Mode
To generate embeddings for an entire dataset:
- Make sure Test mode is disabled.
- Ensure you have a file loaded from the "Load File" tab. The selected "Column X" (containing SMILES) will be used.
- Select an embedder from the dropdown menu.
- Click the "Run" button to start the process.
- The application will process the entire file and save the resulting embeddings as a
.npyfile.
3. Results
After the embedding process is complete, a summary of the results will be displayed, including:
- The dimensions of the generated embedding matrix.
- The number of any invalid SMILES that could not be processed.
- A link to a file containing the invalid SMILES for your review.
Next Steps
Once you have generated the molecular embeddings, you are ready to use them for:
- Applying Dimensionality Reduction to visualize the chemical space.
- Training a Machine Learning Model to predict molecular properties.